-Search query
-Search result
Showing 1 - 50 of 51 items for (author: noel & ea)

EMDB-16493: 
cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117
Method: single particle / : Baquero E, Molinos-Albert L, Mouquet H

EMDB-35326: 
Cryo-EM reconstruction of PBCV-1 capsid block 1
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35490: 
Cryo-EM reconstruction of PBCV-1 capsid block 2
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35585: 
Cryo-EM reconstruction of PBCV-1 capsid block 3
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35586: 
Cryo-EM reconstruction of PBCV-1 capsid block 4
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35589: 
Cryo-EM reconstruction of PBCV-1 capsid block 5
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35593: 
Cryo-EM reconstruction of PBCV-1 capsid block 6
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35594: 
Cryo-EM reconstruction of PBCV-1 capsid block 8
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35644: 
Cryo-EM reconstruction of PBCV-1 capsid block 9
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35645: 
Cryo-EM reconstruction of PBCV-1 capsid block 7
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35646: 
Cryo-EM reconstruction of PBCV-1 capsid block 10
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35647: 
Cryo-EM reconstruction of PBCV-1 capsid block 11
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35648: 
Cryo-EM reconstruction of PBCV-1 capsid block 12
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35650: 
Cryo-EM reconstruction of PBCV-1 capsid block 13
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35656: 
Cryo-EM reconstruction of PBCV-1 capsid block 14
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35659: 
Cryo-EM reconstruction of PBCV-1 capsid block 15
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35695: 
Cryo-EM reconstruction of PBCV-1 capsid block 16
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35954: 
Cryo-EM reconstruction of PBCV-1 capsid block 17
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35955: 
Cryo-EM reconstruction of PBCV-1 capsid block 18
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35956: 
Cryo-EM reconstruction of PBCV-1 capsid block 19
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35957: 
Cryo-EM reconstruction of PBCV-1 capsid block 20
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35958: 
Cryo-EM reconstruction of PBCV-1 capsid block 21
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35959: 
Cryo-EM reconstruction of PBCV-1 capsid block 22
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-35960: 
Cryo-EM reconstruction of PBCV-1 capsid block 23
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q
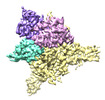
EMDB-27898: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
Method: single particle / : Johnson ZL, Wasilko DJ, Ammirati M, Chang JS, Han S, Wu H
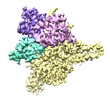
EMDB-27899: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA
Method: single particle / : Wasilko DJ, Johnson ZL, Ammirati M, Chang JS, Han S, Wu H

PDB-8e4y: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
Method: single particle / : Johnson ZL, Wasilko DJ, Ammirati M, Chang JS, Han S, Wu H

PDB-8e50: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA
Method: single particle / : Wasilko DJ, Johnson ZL, Ammirati M, Chang JS, Han S, Wu H

EMDB-34438: 
Near-atomic structure of five-fold averaged PBCV-1 capsid
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-34439: 
Five-fold averaged cryo-EM reconstruction of PBCV-1 before applying the "block-based" reconstruction method
Method: single particle / : Shao Q, Agarkova IV, Noel EA, Dunigan DD, Liu Y, Wang A, Guo M, Xie L, Zhao X, Rossmann MG, Van Etten JL, Klose T, Fang Q

EMDB-27203: 
IMM20190 Fab complex with SARS-CoV-2 Spike Trimer
Method: single particle / : Nikitin PA, DiMuzio JD, Dowling JP, Patel NB, Bingaman-Steele JL, Heimbach BC, Henriquez N, Nicolescu C, Polley A, Sikorski EL, Howanski RJ, Nath M, Shukla H, Scheaffer SM, Finn JP, Liang LF, Smith T, Storm N, McKay LGA, Johnson RI, Malsick LE, Honko AN, Griffiths A, Diamond MS, Sarma P, Geising DH, Morin MJ, Robinson MK

EMDB-27192: 
IMM20253 Fab complex with Trimer Spike protein of SARS-CoV-2 virus
Method: single particle / : Nikitin PA, DiMuzio JD, Dowling JP, Patel NB, Bingaman-Steele JL, Heimbach BC, Henriquez N, Nicolescu C, Polley A, Sikorski EL, Howanski RJ, Nath M, Shukla H, Scheaffer SM, Finn JP, Liang LF, Smith T, Storm N, McKay LGA, Johnson RI, Malsick LE, Honko AN, Griffiths A, Diamond MS, Sarma P, Geising DH, Morin MJ, Robinson MK
EMDB-27193: 
IMM20253 Fab complex with Spike monomer
Method: single particle / : Nikitin PA, DiMuzio JD, Dowling JP, Patel NB, Bingaman-Steele JL, Heimbach BC, Henriquez N, Nicolescu C, Polley A, Sikorski EL, Howanski RJ, Nath M, Shukla H, Scheaffer SM, Finn JP, Liang LF, Smith T, Storm N, McKay LGA, Johnson RI, Malsick LE, Honko AN, Griffiths A, Diamond MS, Sarma P, Geising DH, Morin MJ, Robinson MK
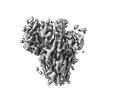
EMDB-27204: 
IMM20184 Fab complex with SARS-CoV-2 Spike Trimer
Method: single particle / : Nikitin PA, DiMuzio JD, Dowling JP, Patel NB, Bingaman-Steele JL, Heimbach BC, Henriquez N, Nicolescu C, Polley A, Sikorski EL, Howanski RJ, Nath M, Shukla H, Scheaffer SM, Finn JP, Liang LF, Smith T, Storm N, McKay LGA, Johnson RI, Malsick LE, Honko AN, Griffiths A, Diamond MS, Sarma P, Geising DH, Morin MJ, Robinson MK

EMDB-27081: 
IMM20184 and IMM20253 Fabs in ternary complex with SARS-CoV-2 receptor binding domain
Method: single particle / : Robinson MK, Nikitin PN

EMDB-23118: 
Orexin Receptor 2 (OX2R) in Complex with G Protein and Natural Peptide-Agonist Orexin B (OxB)
Method: single particle / : Hong C, Byrne NJ, Zamlynny B, Tummala S, Xiao L, Shipman JM, Partridge AT, Minnick C, Breslin MJ, Rudd MT, Stachel SJ, Rada VL, Kern JC, Armacost KA, Hollingsworth SA, O'Brien JA, Hall DL, McDonald TP, Strickland C, Brooun A, Soisson SM, Hollenstein K

EMDB-23119: 
Orexin Receptor 2 (OX2R) in Complex with G Protein and Small-Molecule Agonist Compound 1
Method: single particle / : Hong C, Byrne NJ, Zamlynny B, Tummala S, Xiao L, Shipman JM, Partridge AT, Minnick C, Breslin MJ, Rudd MT, Stachel SJ, Rada VL, Kern JC, Armacost KA, Hollingsworth SA, O'Brien JA, Hall DL, McDonald TP, Strickland C, Brooun A, Soisson SM, Hollenstein K

PDB-7l1u: 
Orexin Receptor 2 (OX2R) in Complex with G Protein and Natural Peptide-Agonist Orexin B (OxB)
Method: single particle / : Hong C, Byrne NJ, Zamlynny B, Tummala S, Xiao L, Shipman JM, Partridge AT, Minnick C, Breslin MJ, Rudd MT, Stachel SJ, Rada VL, Kern JC, Armacost KA, Hollingsworth SA, O'Brien JA, Hall DL, McDonald TP, Strickland C, Brooun A, Soisson SM, Hollenstein K

PDB-7l1v: 
Orexin Receptor 2 (OX2R) in Complex with G Protein and Small-Molecule Agonist Compound 1
Method: single particle / : Hong C, Byrne NJ, Zamlynny B, Tummala S, Xiao L, Shipman JM, Partridge AT, Minnick C, Breslin MJ, Rudd MT, Stachel SJ, Rada VL, Kern JC, Armacost KA, Hollingsworth SA, O'Brien JA, Hall DL, McDonald TP, Strickland C, Brooun A, Soisson SM, Hollenstein K

EMDB-4584: 
Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel J, Wollweber F, Pfitzner A, Muehleip A, Sanchez R, Kudryashev M, Chiaruttin N, Lilie H, Schleger J, Rosenbaum E, Hessenberger M, Matthaeus C, Noe F, Roux A, van der Laan M, Kuehlbrandt W, Daumke O
EMDB-10062: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O
EMDB-10063: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-10064: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-10065: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzt: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O

PDB-6rzu: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzv: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzw: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-3896: 
Cryo-EM Structure of Saccharomyces cerevisiae Target of Rapamycin Complex 2
Method: single particle / : Karuppasamy M, Kusmider B, Oliveira TM, Gaubitz C, Prouteau M, Loewith R, Schaffitzel C

PDB-6emk: 
Cryo-EM Structure of Saccharomyces cerevisiae Target of Rapamycin Complex 2
Method: single particle / : Karuppasamy M, Kusmider B, Oliveira TM, Gaubitz C, Prouteau M, Loewith R, Schaffitzel C
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
